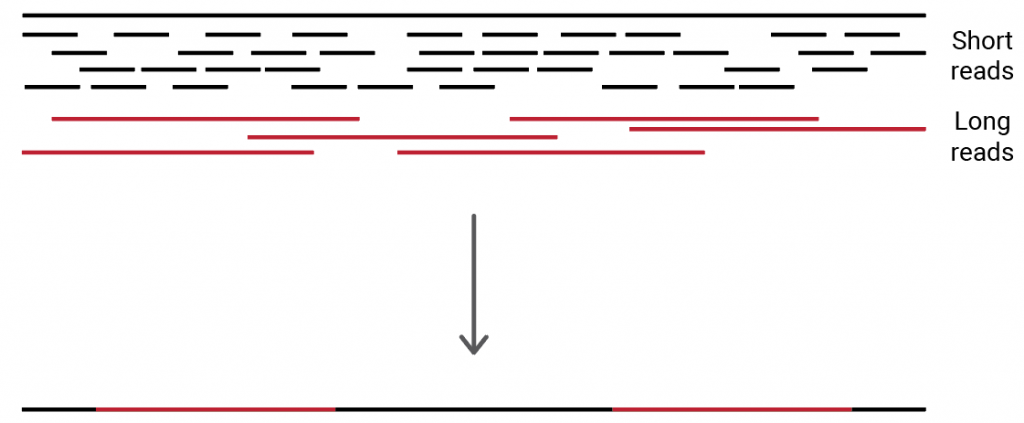
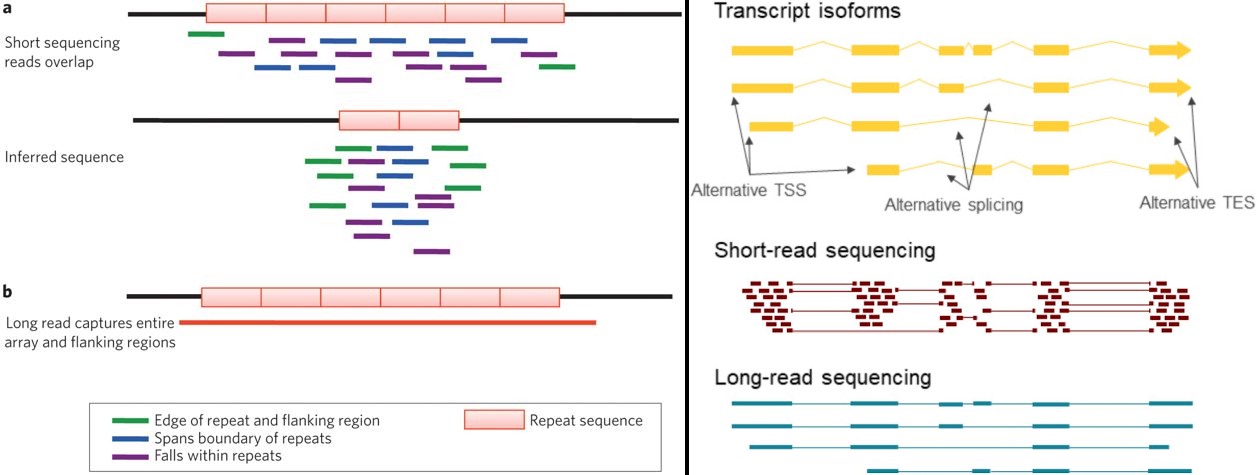
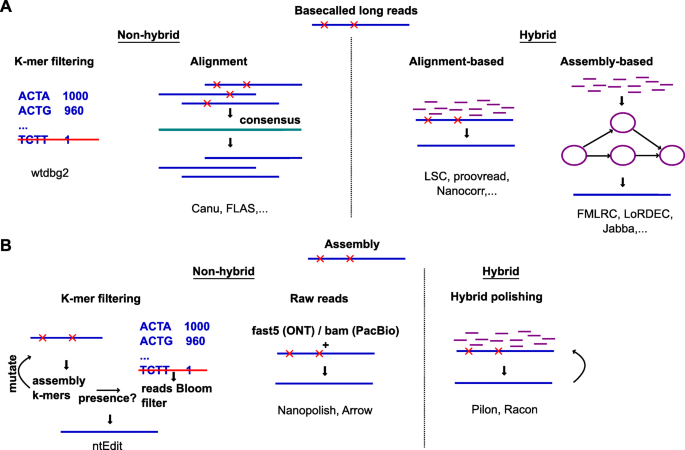
Researchers use a combination of third generation long-read RNA sequencing technology and short-read RNAseq to characterize the splicing landscape of prostate cancer | RNA-Seq Blog
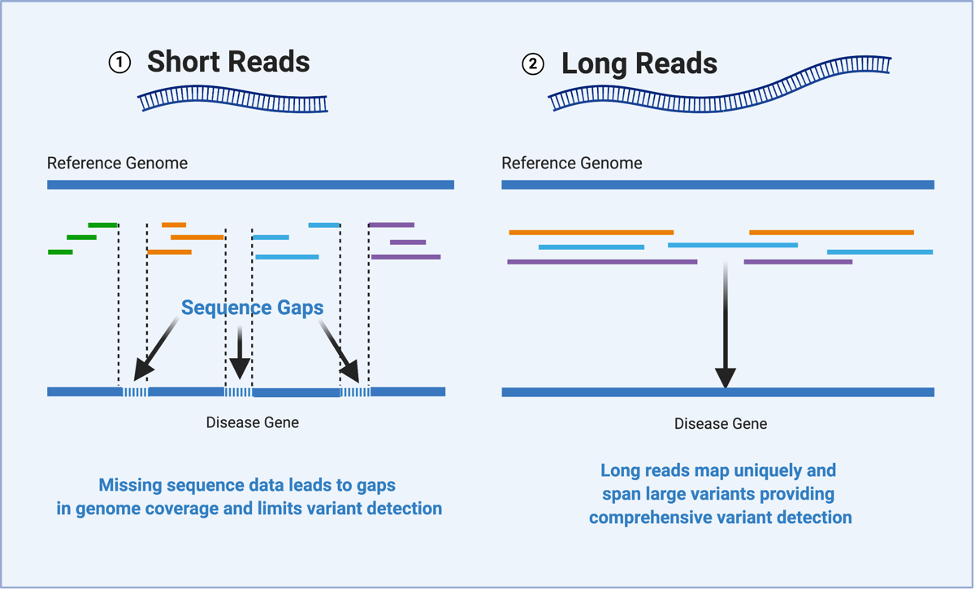
HudsonAlpha researchers use highly accurate long-read sequencing technology to help diagnose rare disease – HudsonAlpha Institute for Biotechnology












![PDF] Long reads: their purpose and place | Semantic Scholar PDF] Long reads: their purpose and place | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/7ac7a62031438eea3b2cd8343147e609d4b5ea85/2-Table1-1.png)